




The GEATbx is build around a layer model. Figure 5-1 gives a first overview of the layers used for the GEATbx.
Fig. 5-1: Layer model of the GEATbx
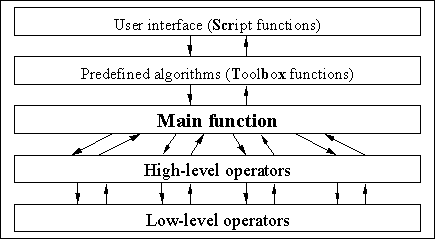
Central point is the Main function. This function is called from the user interface (Script functions). A middle layer (Toolbox functions) defines default parameters for a number of different evolutionary algorithms. The Main function calls all necessary evolutionary operators. The call of evolutionary operators is done by a high-level layer calling the low-level layer. Additionally, the Main function performs nearly all the data management and result collection.
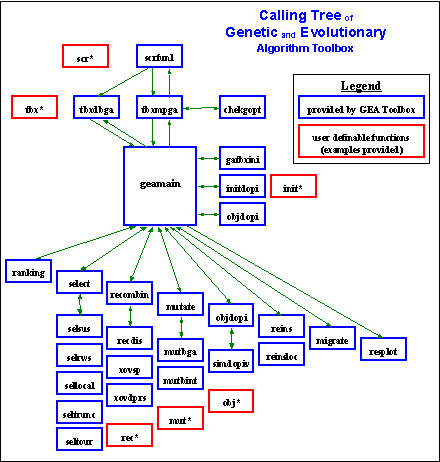
Figure 5-1 shows the full structure of the GEATbx using the names of the corresponding m-functions. Even in this detailed view the layer model is visible. Center of the toolbox is the Main function geamain. The Main function is called by the Script functions via the Toolbox functions (scr* and tbx*). The Main function calls the high-level operators. The high-level operators call the low-level operators. This layer model not only provides a good overview of the structure of the GEATbx. It makes the extension of the toolbox by new operators or special functions straightforward.
5.1 Startup script |
At the beginning there is a high-level script for defining some parameters and the application specific details. These scripts are called "scr*.m".
Script functions serve at the same time as demo functions. Inside a script function every parameter can be preset. Thus, all things necessary to implement special demonstrations are provided by the script functions.
Fig. 5-1: Calling tree of Genetic and Evolutionary Algorithm Toolbox (GEATbx)
5.2 Predefined algorithms |
These scripts call toolbox functions defining an evolutionary algorithm. These functions are called "tbx*.m".
By using one of these functions all appropriate parameters are defined. The remaining parameters are set to default values by chekgopt, checking the correct values at the same time.
5.3 Evolutionary Algorithm - Main function |
The tbx-functions call the working horse of the GEATbx:
Here all the administrative work is done: resolve parameter values, map parameter values to function names (gatbxini), initialize population, run the evolutionary algorithm and display and save results.
The initialization is done by:
As soon as the population of individuals is initialized and evaluated, the evolutionary algorithm starts. For as many generations as necessary/defined, new populations are produced and evaluated. All functions of the evolutionary algorithm are called via high-level functions, thus supporting the multi-population (distributed) concept.
Inside the high-level functions the appropriate functions are called.
For fitness assignment by ranking the toolbox provides 1 function, no low-level functions are used:
For selection a high-level function is provided:
By parameter a low-level selection function can be chosen, that will be called for every subpopulation inside the high-level function:
For recombination/crossover a high-level function is provided:
By parameter low-level recombination/crossover functions can be chosen which will be called for the subpopulations inside the high-level function. The term recombination is used for real valued variables, crossover for binary valued variables (historical reasons).
Recombination:
Crossover:
All crossover functions call the lowest level function xovmp (multi point crossover), which provides all the functionality.
For mutation a high-level function is provided:
Depending on the variable representation one of the low-level mutation functions will be called for every subpopulation:
Real values:
Binary values:
For real value representation a number of additional mutation functions are provided. These functions use the mutation operator of evolutionary strategies. Thus, an adaptation of strategy parameters takes place.
Evolutionary strategy mutation operators:
The toolbox provides many examples for objective functions. These functions are called "obj*.m". All functions use the same calling syntax.
Standard evolutionary algorithm test functions with a free definable dimension using the real value representation are provided in:
Binary value representation is used in:
Dynamic optimization is implemented in:
Standard optimization test functions with 2 independent variables (dimension = 2) are provided in:
See M-functions - Index (part of online documentation) for all functions or Examples of objective functions (part of online documentation) for a mathematical description.
It is possible to pass up to 10 parameters to every objective function.
Instead of writing an objective function file and passing the name of the function, the objective function can be passed directly in a string to the main function as well. However, it is recommended to use one of the provided objective functions as a template and pass over only the file name.
For reinsertion the toolbox provides a high-level function:
Depending on the used population model (global/regional or local), a corresponding low-level reinsertion function is called from inside the high-level reinsertion function.
Low-level reinsertion functions:
For migration the toolbox provides 1 function, no low-level functions are used:
The visualization of the results is implemented on 3 different levels:
5.4 Utility functions |
Throughout the toolbox low-level utility functions are used. They will be useful for everyday work as well: